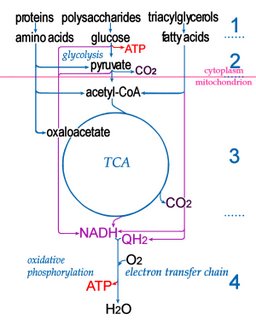
Electron transport chainsElectron transport chains (electron transfer chains) are biochemical reaction sequences that ultimately utilize ATP synthase to produce
ATP, the energy currency of life.
Only two sources of energy are available to living organisms: oxidation-reduction (redox*) reactions and photic energy (
photosynthesis). Chemotrophic organisms employ redox reactions to produce
ATP. Phototrophic organisms employ light as their initial energy source. Both chemotrophs and phototrophs utilize electron transport chains to convert energy into ATP.
Table
Electron Transport vs Oxidative Phosphorylation :
Oxidation involves loss of electrons, and
reduction involves gain of electrons (mnemonic 'oilrig').

Redox reactions
*Because electrons may not be transfered in redox reactions, oxidation is better defined as an increase in oxidation number, and reduction as a decrease in oxidation number. Oxidants are typically highly electronegative molecules. Redox reactions are chemical reactions in which electrons are transferred from a donor molecule to an acceptor molecule. The reductant (electropositive metal, hydride) transfers electrons to the electronegative oxidant. The Gibbs free energy of the reactants compared to the products provides the chemical potential energy for redox reactions.
The Gibbs free energy is the energy available (“free”) to do work. Any reaction that decreases the overall Gibbs free energy of a system (reactants → products) will proceed spontaneously with release of that energy. The transfer of electrons from a high-energy donor molecule to a lower-energy acceptor molecule can be spatially separated into a series of intermediate redox reactions within an electron transport chain. The spatial separation permits biological control of the reactions.
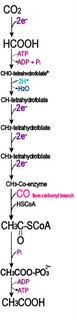
Many metabolic reactions involve the storage and release of biological energy by means of redox reactions. Photosynthesis involves the reduction of CO2 into sugars along with the oxidation of H2O into O2. The carbon fixing, light-independent reactions of photosynthesis employ the Calvin cycle ( C-3) in most organisms, though carbon fixation may also employ the 3-hydroxypropionate cycle, the reductive acetyl CoA pathway, or the reverse tricarboxyclic acid cycle. The reverse reaction, respiration, oxidizes sugars to produce CO2 + H2O.
In intermediate steps, reduction of oxygen accompanies the employment of reduced carbon compounds to reduce nicotinamide adenine dinucleotide (NAD+ → NADH), which then contributes to the creation of a proton gradient, which in turn drives the synthesis by ATP synthase of adenosine triphosphate (ADP + Pi → ATP).
Within animal cells, mitochondria perform similar functions to the photophosphorylation reactions within the chlorosomes of prokaryotes, the phycobilin studded thylakoid membranes of Cyanobacteria, or the chloroplasts of plant cells.
The term redox state is often used to describe the balance of NAD+/NADH and NADP+/NADPH in a biological system such as a cell or organ. The redox state is reflected in the balance of several sets of metabolites such as lactate and pyruvate, and beta-hydroxybutyrate and acetoacetate whose interconversion is dependent on these ratios. An abnormal redox state can develop in a variety of deleterious situations, such as hypoxia, shock, and sepsis.
Within mitochondria, a complex series of transmembrane proteins perform the overall reaction of oxidative phosporylation:
NADH → Complex I → Q → Complex III → cytochrome c → Complex IV → O2
.................................. ↑
...........................Complex II
Within chloroplasts, the membrane-bound pigment-protein complexes of photosystem I perform cyclic photophosphorylation, and the sequence of photosystem I & photosystem II perform noncyclic photophosphorylation (image - Z-scheme).
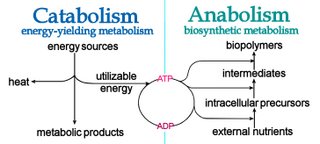 Anabolism, as in ‘anabolic steroids’, refers to those metabolic processes that utilize energy to biosynthesize complex molecules and to generate growth.
Anabolism, as in ‘anabolic steroids’, refers to those metabolic processes that utilize energy to biosynthesize complex molecules and to generate growth.